| Primary Citation | Enhancing RBD exposure and S1 shedding by an extremely conserved SARS-CoV-2 NTD epitope. Zhu Q, Liu P, Liu S, Yue C, Wang X Signal Transduct Target Ther (2024) PubMed DOI show all entries for this citation |
|---|
| Funding Source | Not funded |
|---|
| Status | Deposited | Released | Last Update |
|---|
| REL | 2024-01-30 | 2024-09-04 | 2024-11-06 |
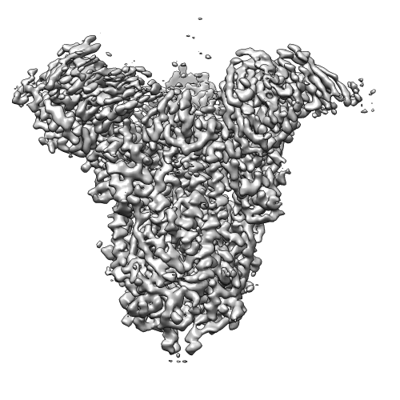
Name: BA.2.86 S-trimer in complex with Nab XGv280SpecimenType: particle MW: 0.489 MDa pH: 7.4
| Component Types | complex |
|---|
| Component Names | BA.2.86 S-trimer in complex with Nab XGv280 |
|---|
| Molecular Types | ligand, protein |
|---|
| Molecule Names | - 2-acetamido-2-deoxy-beta-D-glucopyranose
- Spike glycoprotein
- XGv280 Heavy chain
- XGv280 Light chain
|
|---|
| Sources | Homo sapiens ncbi:9606 Severe acute respiratory syndrome... ncbi:2697049 |
|---|
Ligand IDs: NAG
| Specimen Preparation | vitrification |
|---|
| Microscope | FEI TITAN (60 kV) |
|---|
| Detector | GATAN K2 SUMMIT (4k x 4k) |
|---|
| Electron Dose | 60 (e/sq. Å) |
|---|
| Number of Particles | 109404 |
|---|
| Map File Size | 131 Mbytes |
|---|
| Voxel Dimensions (Å) | 1.04 x 1.04 x 1.04 |
|---|
| Map Dimensions (voxels) | 320 x 320 x 320 |
|---|
| Map Data Type | IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) |
|---|
| Density Statistics | Min -1.353Max 2.389Avg 0Std 0.068 |
|---|
| Recommended Contour Level | 0.5 (7.3 σ) |
|---|
Mol*3DViewer Map Visual Analysis @ EMDBCurrent: files.wwpdb.org (USA)
| View EMDB map entry EMD-38915: |    |
|---|
| View associated model PDB 8y4c: |    |
|---|