Improved 3D Viewing for EM Maps and Models
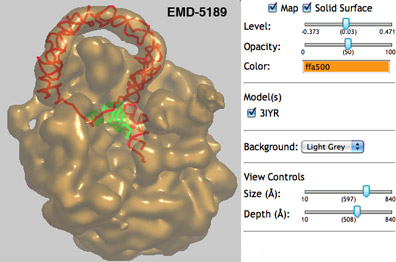
We are pleased to announce improved 3D web-based viewing of maps and associated coordinate models from our EMDB atlas pages.
The molecular visualization program OpenAstexViewer has been adapted by our team to display EM maps and their associated coordinate models. Clicking on the "Launch Astex Viewer" button on an atlas visualization page will bring up the EMDB map and any fitted PDB models. The map can be displayed either as a solid surface or mesh, and can be adjusted for contour level, opacity, and color. The display can be manipulated using sliders for size and depth as well as mouse drag and click actions. Map and model display can be turned on or off using check-boxes. Models are displayed as backbone traces, including any symmetry information available in the PDB entry. Residues can be identified by selecting an atom in the model.
You can try launching astexviewer for the example shown here, a tmRNA-SmpB bacterial ribosome complex, from our UK and USA partner site visualization pages.
We welcome your feedback on the updated viewer and all other services. Please send your comments and suggestions to help@emdataresource.org.
